Written by Nascent Transcript writer Kathryn Vaillancourt
The invention of the CRISPR/Cas9 system for gene editing has revolutionized the way we study molecular genetics. Labs all over the world are adapting this system to delete, mutate, or replace segments of DNA to study the effects on cell functioning and disease, but what if your research question calls for something a little different?
Using a catalytically ‘dead’ version of the enzyme, the CRISPR/Cas9 family tree continues to grow. Instead of “cut-and-paste”, the “dead” version of the Cas9 (dCas9) endonuclease has been mutated to perform a massive range of functions in mammalian cells. Although some require other factors to be transfected with the dCas9 plasmid, they all rely on the now famous CRISPR guide RNA (gRNA) system.
Transcriptional Regulators
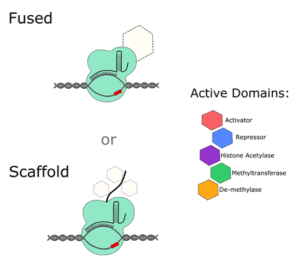
If you want to alter the expression of a gene without editing any of its regulatory sequences, take a look at dCas9 activators and repressors. Most systems that are available (Addgene has over 100 mammalian dCas9 activators alone), are either fused to an active domain of a regulatory protein, or to a scaffold that recruits and amplifies the effect of other regulatory factors within the cell.
One of the earliest CRISPR activator systems paired dCas9 with 4 copies of the 16 amino acid viral protein (VP64) transcription factor (TF). Since then, researchers have ramped up gene expression using other TFs and have even fused multiple TFs together to try and get the most out of each transfection. For example, the dCas9-VPR system contains the active domains of VP64, p65 and RTA together. A study comparing multiple activators suggests that, if you really need maximum expression in mammalian cells, a scaffold system is your best bet, either at the gRNA level or with a Suntag fused to the dCas9 protein.
On the other hand, you can interfere with gene expression by using a dCas9 repressor plasmid. Although there are slightly fewer of these systems to choose from, they’re designed in a similar way to activator systems. They often contain transcriptional repressors, like Krüppel associated box proteins (KRAB) and gRNAs directed to promoter or enhancer sequences.
Epigenetic Editors
Of course, there is more to gene expression than the presence or absence of transcription factors; epigenetic mechanisms are important regulators of gene activity, and the world of dCas9 has multiple offerings in this field as well. At the DNA level, there are multiple plasmids available that can increase methylation through the de novo methyltransferase (DNMT3) or reduce methylation through the ten eleven methylcytosine dioxygenase (TET1).
Histone post-translational modifications add another layer of complexity and have also started to be targets of dCas9 systems. To date, dCas9 plasmids have been designed to selectively add acetyl groups to histone tails, via histone acetyltransferase activity, or remove methyl groups using lysine-specific histone demethylase 1A (LSD1). As we continue to try and fully understand the function of the over 4,000 combinations of histone modifications in the human genome, our options will undoubtedly increase.
And More!
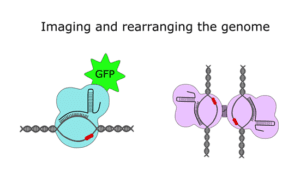
If that hasn’t scratched your Cas9 itch, there’s another branch of research that has come up with creative ways to use the protein that go beyond (epi)genome editing. In 2013, researchers combined dCas9 with green fluorescent protein (GFP) to map the location of specific genomic segments inside cells, and more recently, CRISPR-based DNA imaging has been expanded to visualize multiple loci in a rainbow of colors. The CRISPR/Cas9 system has also been adapted to recruit non-coding RNAs to particular regions of the genome and to bring distant genomic loci together through chromatin looping.
With new iterations being constantly released, like new modular systems that use nanobodies to recruit any tagged protein, it will be exciting to see what the next generation of dCas9 technologies will bring!
